August 11, 2016
Super-resolution 3-D microscopy images cells in unprecedented detail
 Using a new 3-D super-resolution instrument, researchers captured this visualization of a "primary cilium," the antenna of the cell. (Photo courtesy of Huang et al./Cell)
Download image
Using a new 3-D super-resolution instrument, researchers captured this visualization of a "primary cilium," the antenna of the cell. (Photo courtesy of Huang et al./Cell)
Download image
WEST LAFAYETTE, Ind. – A new ultra-high resolution "nanoscope" is capable of taking 3-D images of an entire cell and its cellular constituents in unprecedented detail, an advance that could reveal biological phenomena never before seen and bring new medical insights.
The work was carried out by researchers at Yale University, Purdue University, the University of Cambridge, the Jackson Laboratory, Howard Hughes Medical Institute and the University of Oxford.
The technology incorporates several innovations in fluorescence microscopy and super-resolution microscopy and harnesses the same kind of "adaptive optics" technology used in astronomy - deformable mirrors that change shape to compensate for light distortion. In astronomy the deformable mirrors are used to compensate for atmospheric distortion to yield clear images of celestial objects. Deformable mirrors also can be used to counteract the distortion caused when light passes through biological tissue.
Super-resolution fluorescence microscopy earned its developers the 2014 Nobel Prize in chemistry and has become an important tool in cell biology research. Its use, however, has been limited because of challenges in resolving features deep below the surface of samples.
Researchers have now solved that problem with the development of the new system, called whole-cell 4Pi single-molecule switching nanoscopy (W-4PiSMSN), said Fang Huang, an assistant professor of biomedical engineering at Purdue.
Findings are detailed in a research paper appearing Thursday (Aug. 11) in the journal Cell. The paper's co-lead authors are Huang and Yale postdoctoral research associate George Sirinakis. The corresponding author is Joerg Bewersdorf, a Yale associate professor of cell biology and biomedical engineering. (A YouTube video is available at https://youtu.be/hdnFTnvJxjM)
The technology was demonstrated by imaging cellular components, including synaptonemal complexes, which link chromosomes together; mitochondria and the endoplasmic reticulum, which are critical for cellular functions; the cilia, which protrude from cells like tiny antennas; and bacteriophages, which are viruses about 50 nanometers in diameter that infect bacteria.
The new system advances the original super-resolution fluorescence microscopy by incorporating the deformable mirrors into a microscope using two objectives, one above and one below the sample, and introducing a set of new algorithms to pinpoint molecular positions of proteins deep inside cells.
The system allows researchers to resolve details far smaller than the wavelength of light, representing a powerful and versatile new laboratory tool, Huang said.
"The wave nature of light restricts the resolution of conventional light microscopy to about 200 nanometers, making details of subcellular structures and protein assemblies unresolvable," Huang said.
The new system, however, allows imaging of cellular constituents in 3-D at 10- to 20-nanometer resolution throughout entire mammalian cells, powerful enough to reconstruct the fine features of viruses. Until now, resolving such fine details was only possible using electron microscopy, which requires samples to be treated, killing the cells.
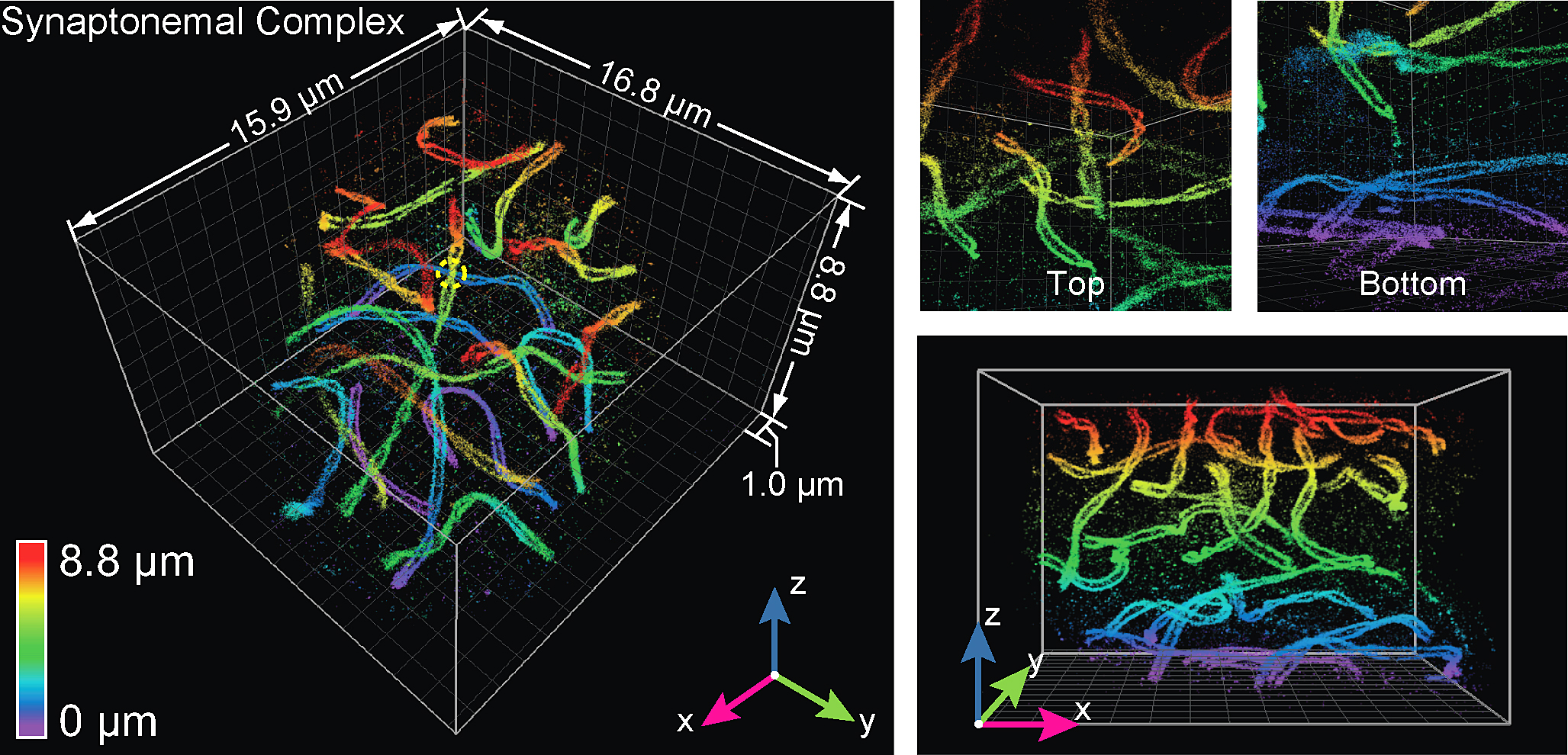 Here, the technology was used to visualize a mouse spermatocyte, revealing with unprecedented clarity the "twisting paired lateral elements" of synaptonemal complexes, which link chromosomes together. (Photo courtesy of Huang et al./Cell)
Download image
Here, the technology was used to visualize a mouse spermatocyte, revealing with unprecedented clarity the "twisting paired lateral elements" of synaptonemal complexes, which link chromosomes together. (Photo courtesy of Huang et al./Cell)
Download image
"One goal is to further push the envelope in the direction of live-cell and tissue imaging, two major roadblocks of modern super-resolution techniques," Huang said, "and therefore allow visualization of cellular functions live in their physiological conditions at the nanoscale."
Molecules inside cells and in structures called organelles can be tagged with either "photo-switchable" fluorescent proteins or organic dyes that are able to glow when exposed to a small amount of ultraviolet light.
"These special fluorescent tags (fluorophores) have two states, on and off," Huang said. "And you can control the on and off states by shining light on these molecules. The concept of single molecule switching nanoscopy is to stochastically switch molecules on and off at different time frames, pinpoint the exact locations of single molecules and reconstruct the cellular constituents at super resolution."
The super-resolved images are reconstructed from the positions of thousands to millions of single molecules.
"We are interested in using our developments to study the cytokinetic apparatus, a core machinery during cell division," Huang Said.
A complete list of the paper's authors is available in the abstract below.
This work was primarily supported by a grant from the Wellcome Trust (095927/ A/11/Z).
Writer: Emil Venere, 765-494-4709, venere@purdue.edu
Source: Fang Huang, 765-494-6216, fanghuang@purdue.edu
Note to Journalists: A copy of the research paper is available from Emil Venere, Purdue News Service, at 765-494-4709, venere@purdue.edu. A YouTube video is available at https://youtu.be/hdnFTnvJxjM and other videos and graphics are available on Google Drive at https://goo.gl/PYmg7S.
ABSTRACT
Ultra-High Resolution 3D Imaging of Whole Cells
Fang Huang,1,2,14 George Sirinakis,1,3,14 Edward S. Allgeyer,1,3 Lena K. Schroeder,1 Whitney C. Duim,1,4, Emil B. Kromann,1,5 Thomy Phan,1 Felix E. Rivera-Molina,1 Jordan R. Myers,1 Irnov Irnov,6,7 Mark Lessard,8 Yongdeng Zhang,1 Mary Ann Handel,8 Christine Jacobs-Wagner,6,7,9,10 C. Patrick Lusk,1 James E. Rothman,1,11 Derek Toomre,1,11 Martin J. Booth,12,13 and Joerg Bewersdorf1,5,11,*
1Department of Cell Biology, School of Medicine, Yale University, New Haven, CT 06520, USA
2Weldon School of Biomedical Engineering, Purdue University, West Lafayette, IN 47907, USA
3The Gurdon Institute, University of Cambridge, Cambridge CB2 1QN, UK
4Department of Chemistry, Harvey Mudd College, Claremont, CA 91711, USA
5Department of Biomedical Engineering, Yale University, CT 06520, USA
6Microbial Sciences Institute, Yale University, West Haven, CT 06516, USA
7Department of Molecular, Cellular and Developmental Biology, Yale University, New Haven, CT
8The Jackson Laboratory, Bar Harbor, ME 04609, USA
9Howard Hughes Medical Institute, Yale University, New Haven, CT 06520, USA
10Department of Microbial Pathogenesis, Yale School of Medicine, New Haven, CT 06520, USA
11Nanobiology Institute, Yale University, West Haven, CT 06516, USA
12Department of Engineering Science, University of Oxford, Oxford OX1 3PJ, UK
13Centre for Neural Circuits and Behaviour, University of Oxford, Oxford OX1 3SR, UK
14Co-first author
*Correspondence: joerg.bewersdorf@yale.edu
Fluorescence nanoscopy, or super-resolution microscopy, has become an important tool in cell biological research. However, because of its usually inferior resolution in the depth direction (50-80 nm) and rapidly deteriorating resolution in thick samples, its practical biological application has been effectively limited to two dimensions and thin samples. Here, we present the development of whole-cell 4Pi single-molecule switching nanoscopy (W-4PiSMSN), an optical nanoscope that allows imaging of three-dimensional (3D) structures at 10- to 20-nm resolution throughout entire mammalian cells. We demonstrate the wide applicability of W-4PiSMSN across diverse research fields by imaging complex molecular architectures ranging from bacteriophages to nuclear pores, cilia, and synaptonemal complexes in large 3-D cellular volumes.

