June 28, 2018
Mapping the brain with data science
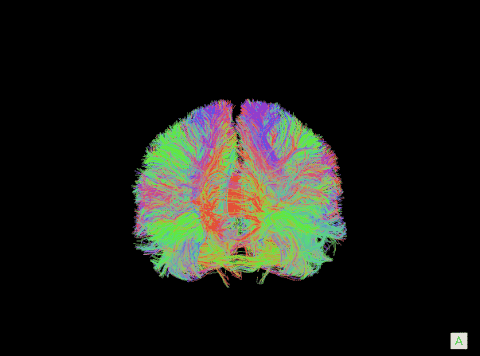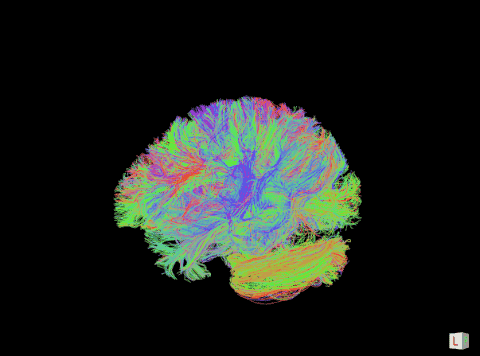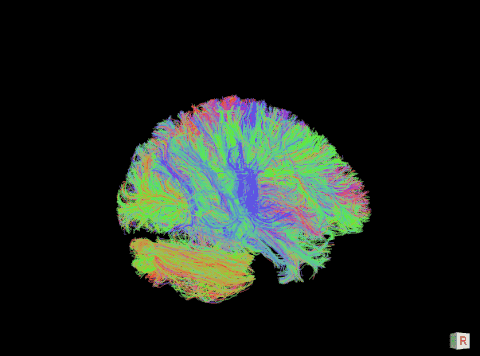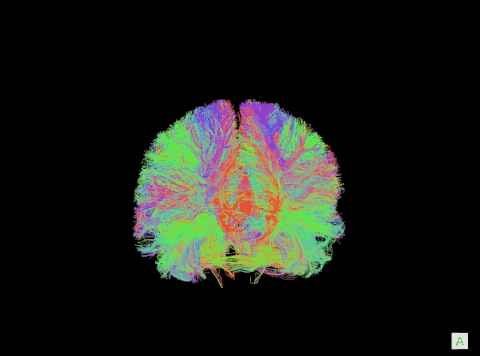
A visualization of the human connectome. (Purdue University image)
Visualizing neural connections gives scientists new insight into neurological diseases
WEST LAFAYETTE, Ind. — Patients with dementia and other neural diseases show physical symptoms such as stumbling and confusion, but identifying the problem isn’t as simple as taking an X-ray. A group of researchers at Purdue University are designing data-driven tools that will help clinicians better understand the progression of neurodegenerative diseases by identifying and tracking changes in the brain.
“We’re not to the point where we’re taking X-rays to see if you have a broken bone in your leg, but we’re at least at the stage where we’re saying, ‘Your gait is very funny,’” said Tom Talavage, professor of electrical and computer engineering and biomedical engineering, and a co-investigator for the project. “We can narrow it down to something wrong with your leg, and we can make inferences about what’s wrong with your leg. We can say, ‘You probably have a broken leg because of how you’re walking.’ That’s what we’re really getting at.”
The project is led by Joaquín Goñi, an assistant professor of industrial engineering and biomedical engineering who studies the network of neural connections composing the human brain. This network is called the connectome, the focus of an emerging field of study known as brain connectomics. Brain-imaging techniques, such as diffusion weighted imaging and functional magnetic resonance imaging (fMRI), allow neuroscientists to model and examine the connectome to understand communication between different regions of the brain. This helps them see which parts of the brain are functioning normally - and which regions are not - by observing changes over time.
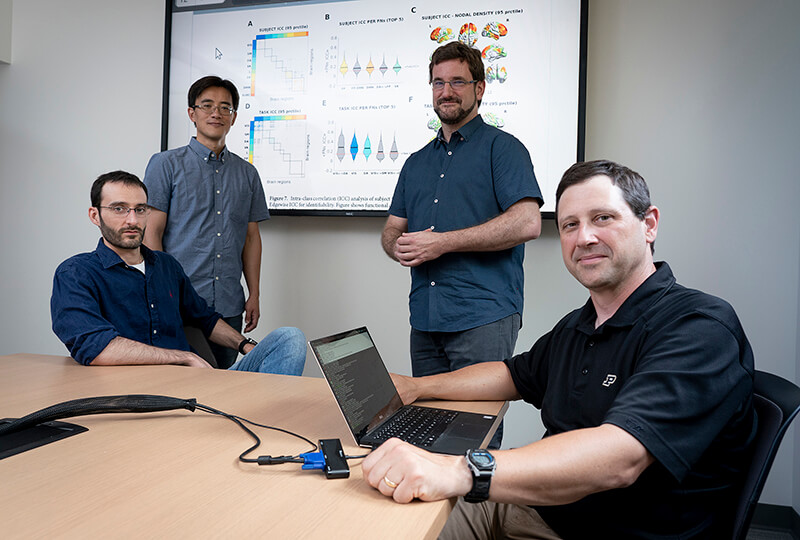 Researchers at Purdue are mapping the neural connections of the human brain to better understand the progression of neurological diseases. From left are Mario Ventresca, Yunjie Tong, Joaquin Goni, and Thomas Talavage. (Purdue University photo)
Download image
Researchers at Purdue are mapping the neural connections of the human brain to better understand the progression of neurological diseases. From left are Mario Ventresca, Yunjie Tong, Joaquin Goni, and Thomas Talavage. (Purdue University photo)
Download image
“What we’re really doing is starting to create the means to see symptoms of neurodegenerative diseases; they become physical in the graphical approaches we apply. It’s very visual,” Talavage said.
The team is using data from the Human Connectome Project, a collection of data sets from different projects focused on the connectome, to research and develop their method. Goñi and postdoctoral researcher Enrico Amico recently published a paper in Nature Scientific Reports using data from the Human Connectome Project to propose a data-driven method for assessing the connectome. Their work demonstrates that individual connectomes are unique enough to be identifiable – and could potentially be used to better understand the differences between people’s individual connectomes and how this relates to their health.
Their project uses brain-imaging data that shows both “structural” and “functional” connectivity – physical connections between different regions of the brain, and the communication between those regions as a subject performs a given task. Making sense of an individual person’s network of connections and understanding their cognitive health depends on images of brain activity collected during these tasks.
“When my brain is performing activities, is it more consistently, uniquely me? Or does the task actually make me look like everybody else?” Talavage said. “The end results indicate that we are more uniquely ourselves even when we’re performing the same task.”
Taking these detailed pictures of the connectome can be a challenge for neuroscientists. Often the neuro-images are affected by visual noise, which makes it more difficult to make sense of the data.
“We want accurate representations of both structural and functional connectivity. Individual estimations of brain connectivity are quite noisy, meaning that they are not necessarily accurate in representing the subject’s characteristics,” said Goñi, who is the principal investigator for the research project. “We know that if we take the average of all those mapping connections, we get fairly good representations of the human connectomes.”
One of the areas their research explores is to what extent the individual’s neural connections resemble each other because of genetics. Goñi and his colleagues are using the Humane Connectome Project’s data from pairs of identical (“monozygotic”) and fraternal (“dizygotic”) twins to see if genetic resemblance plays a role. If their method incorrectly identified one monozygotic twin as their identical counterpart, the team could then correct this mistake and improve the reliability of their results with each test.
“Joaquin is working on a large study with twin data to document whether the twins, who were monozygotic versus dizygotic, looked like one another in terms of brain connectivity -- and of course, they do.” Talavage said. “But they are still able to pull out the individual twin from the other twin. Even two identical twins do not look closer to one another in the end.”
Although the result did not surprise the team, it did reassure them.
“I don’t think it’s particularly shocking,” Talavage said. “But at the same time it is a very important illustration about the subtleties that he can start pulling out. Yes, you look more like your twin than everybody else, but you’re still differentiable.”
Ultimately the team hopes to release a proven tool for assessing the human connectome that will help clinicians track disease progression and neural health.
“In many diseases, it’s not like there is a unique progression of the disease that could be assumed for all the patients,” Goñi said. “Having individual, accurate structural and functional connectomes is crucial for better understanding how the progression is affecting different parts of the brain, different subsequets and to what extent how much these are affected. These have implications in behavioral and neuropsychological evaluations.”
“It’s really just allowing us to look at the overall functioning of the brain in a very concise manner,” Talavage said. “Then we can assess the state of the brain and the trajectory it’s following.”
The team’s project was one of eight that were chosen for Purdue’s Integrative Data Science Initiative after a competition for funding. The initiative will advance Purdue’s data science-related research and education by funding projects that target four focus areas: health care; defense; fundamentals, methods, and algorithms; and ethics, society and policy. The projects were chosen to integrate data science programs with campus-wide courses and curricula, and are hosted by Purdue’s Discovery Park.
The interdisciplinary research team is composed of four faculty members from several engineering departments at Purdue. Other members of the team include Yunjie Tong, assistant professor of biomedical engineering, and Mario Ventresca, assistant professor of industrial engineering. After the conclusion of the two-year funding period with the Integrative Data Science Initiative, the team hopes to work with clinicians on implementing their tool.
“When I came three years ago to Purdue, one of my most ambitious aims was that we get Purdue to be known for our work on brain connectomics,” Goñi said. “I really think that thanks to this project, we have a chance to make that happen. In my name and in the name of all the other team members, we are really grateful for this opportunity and this project. We are super excited to move forward.”
About Discovery Park
Discovery Park is an open laboratory for interdisciplinary collaboration focused on global challenges. Its mission is to accelerate world-changing interdisciplinary research, enrich transformative education and advance the translation of innovation to commercialization activities of faculty, students and staff.
Writer: Sarah Olson, 760-419-9222, seolson@purdue.edu
Sources: Joaquín Goñi, jgonicor@purdue.edu
Tom Talavage, tmt@purdue.edu

