New findings detail how virus prepares to infect cells
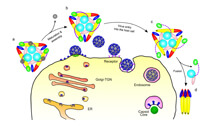
This diagram depicts how the changing arrangement of proteins in a key structure enables viruses to invade and fuse with host cells. (Purdue University Department of Biological Sciences image/Long Li)
WEST LAFAYETTE, Ind. - Researchers have learned the atomic-scale arrangement of proteins in a structure that enables a virus to invade and fuse with host cells, showing precisely how the structure morphs with changing acidity to initiate infection.
Findings from a team at Purdue University showed the protein structure in an acidic environment, and another team from the Pasteur Institute showed the same structure in a neutral environment. When combined, the two studies illustrate what happens to the structure as a virus enters and then prepares to fuse with a host cell, critical steps leading to infection.
"These findings represent a milestone," said Michael Rossmann, Purdue's Hanley Distinguished Professor of Biological Sciences, who is working with Long Li, a postdoctoral researcher in his lab, and Joyce Jose, a postdoctoral researcher in the laboratory of Richard Kuhn, a professor and head of Purdue's Department of Biological Sciences.
The research is aimed at learning precisely how viruses infect humans and other hosts, knowledge that may lead to better vaccines and antiviral drugs, Rossmann said.
Findings from the Purdue and Pasteur Institute studies are detailed in two papers appearing in the journal Nature on Thursday (Dec. 2). The Purdue paper was written by Li, Jose, postdoctoral researcher Ye Xiang, Kuhn and Rossmann.
The researchers studied alphaviruses, a family of viruses that includes eastern equine encephalitis and chikungunya viruses, which are transmitted by mosquitoes and sometimes ticks. The work focused on two "envelope proteins" making up 80 spikelike structures protruding from the outer shell of the viruses.
"The spikes have all the machinery for infecting a cell," Rossmann said.
Researchers have known the structure of envelop protein 1, or E1, for several years. The Purdue researchers have now determined the structure of envelope protein 2 and the precise atomic-scale architecture of the combined E1-E2 complex. Scientists had previously determined general characteristics about E2, such as its location in the protein complex, but they did not know its structure until now.
E2, a receptor-binding protein, enables the virus to initially attach to and enter host cells where the virus encounters an acidic environment that induces changes in the structure of the protein complex. These changes expose a portion of E1 required to fuse the virus with the cell membrane, leading to the formation of a "fusion pore" through which the virus's genetic material is transferred into the host cell. Once infected, the host cell then produces new virus particles.
The researchers learned the shape of E2's three "domains," showing how E2 displaces one of these domains when in an acidic environment, allowing fusion with the cell membrane. The scientists used advanced imaging technologies, including cryoelectron microscopy and X-ray crystallography, to uncover critical structural details about the viruses.
Purdue researchers led by Rossmann and Kuhn have been studying alphaviruses for about 15 years, in work based at Purdue's Markey Center for Structural Biology.
The research is funded by the National Institutes of Health.
Purdue also is leading a team of researchers in a federally funded effort aimed ultimately at developing better vaccines and antiviral drugs against alphaviruses and flaviviruses, a family that includes West Nile and dengue.
Writer: Emil Venere, 765-494-4709, venere@purdue.edu
Sources: Richard J. Kuhn, 765-494-1164, kuhnr@purdue.edu
Michael Rossmann, 765-494-4911, mgr@indiana.bio.purdue.edu
Note to Journalists: Journalists may obtain a copy of the research paper by contacting Nature at press@nature.com or calling 212-726-9231.
ABSTRACT
Structural Changes of Envelope Proteins During Alphavirus Fusion
Long Li, Joyce Jose, Ye Xiang, Richard J. Kuhn, & Michael G. Rossmann
Department of Biological Sciences, Purdue University
Alphaviruses are enveloped RNA viruses that have a diameter of about 700 Å and can be lethal human pathogens1 (Fig. 1). Entry of virus into host cells by endocytosis is controlled by two envelope glycoproteins, E1 and E2. The E2-E1 heterodimers form 80 trimeric spikes on the icosahedral virus surface1,2, 60 with quasi-threefold symmetry and 20 coincident with the icosahedral threefold axes arranged with T=4 quasi-symmetry (Fig. 1a). The E1 glycoprotein has a hydrophobic fusion loop at one end and is responsible for membrane fusion3,4. The E2 protein is responsible for receptor binding5,6 and protects the fusion loop at neutral pH. The lower pH in the endosome induces the virions to undergo an irreversible conformational change in which E2 and E1 dissociate and E1 forms homotrimers, triggering fusion of the viral membrane with the endosomal membrane and then releasing the viral genome into the cytoplasm3,4. Here we report the structure of an alphavirus spike, crystallized at low pH, representing an intermediate in the fusion process and clarifying the maturation process. The trimer of E2-E1 in the crystal structure is similar to the spikes in the neutral pH virus except that the E2 middle region is disordered, exposing the fusion loop. The amino- and carboxy-terminal domains of E2 each form immunoglobulin-like folds, consistent with the receptor attachment properties of E2.
