July 5, 2016
LobeFinder technology quantifies changes in shape-shifting plant cells
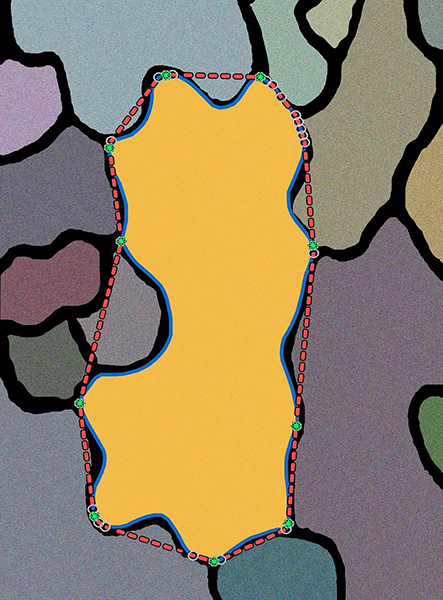 A field pf pavement cells comprising the surface is shown in this graphical image. The cell in the middle has been analyzed with LobeFinder to identify the number and position of lobes reflecting the growth history of the cell. (Purdue Botany and Plant Pathology/Agronomy graphic/Daniel Szymanski)
Download image
A field pf pavement cells comprising the surface is shown in this graphical image. The cell in the middle has been analyzed with LobeFinder to identify the number and position of lobes reflecting the growth history of the cell. (Purdue Botany and Plant Pathology/Agronomy graphic/Daniel Szymanski)
Download image
WEST LAFAYETTE, Ind. - Purdue University researchers have developed an algorithm that quantifies and analyzes shape changes in puzzle piece-shaped plant cells, providing insights into the small-scale processes that control leaf size and crop yield.
The technology could also be adapted to measure boundary shifts in other complex geometric forms, from neuron cells and tumors to shorelines and glaciers.
Researchers Daniel Szymanski and David Umulis led an interdisciplinary team to create LobeFinder, a tool that helps troubleshoot one of plant biologists' trickiest tasks - quantifying changes in the perimeter of pavement cells, highly geometric plant cells that vary greatly in size and shape. Together, pavement cells form a mosaic that drives the expansion of leaves, which act like solar panels, absorbing sunlight and converting it to energy.
Understanding how these cells morph from simple polyhedral shapes into what resemble jigsaw puzzle pieces could enable long-term crop improvement as changes in pavement cells impact leaf area and expansion, key components of yield.
But identifying shape changes in pavement cells has largely been a manual and subjective process, a researcher eyeballing a cell image and characterizing what she thinks she sees.
LobeFinder is designed to remove the guesswork and standardize the data, analyzing cell shapes and providing graphs of cell growth behavior in a matter of seconds per cell.
"Cells are the building blocks of plant tissues and organs. The number, size and shape of cells ultimately control the size, shape and mechanical properties of leaves," said Szymanski, professor of botany and plant pathology and agronomy. "If you want to be able to engineer plant architecture, you need to understand plant processes at multiple scales - how proteins influence the shape of cells and how those cell shape patterns can function collectively to influence plant features such as leaf area and yield."
Pavement cells undergo a complicated process of division and expansion to form a leaf, with varying rates and patterns of growth across a population of cells and even within different regions of a single cell. As the cells grow, they form lobes, angled protrusions that signal cell shape change. If a pentagon were a pavement cell, for example, it would have five lobes - the angles that make up its five points.
Identifying, counting and analyzing lobes helps researchers understand cell shape behavior and scale up this knowledge to characterize how a leaf reaches its size and shape. A change in cell geometry or shape can signal a change within the cell's chemistry or a defect caused by a mutation.
But the manual scoring of lobe number results in great variability in data among individuals. Where one expert sees a lobe, another might not. When results from a panel of experts are pooled, however, the data becomes consistent.
Szymanski and Umulis used one such pool of results to "train" LobeFinder to identify and count lobes, plot cell growth behavior on a graph, and analyze changes in cell boundary over time. They validated LobeFinder's results with lobe data from a panel of plant scientists.
"Subjective scoring of cell expansion has been a major impediment in the field for this cell type," Szymanski said. "LobeFinder is going to give everyone the same answer."
The algorithm can run on a personal computer with MATLAB software.
"We wanted a tool that was simple and objective, easily interpretable for human curators of the data," said Umulis, associate professor of agricultural and biological engineering and biomedical engineering. "LobeFinder offers a really good way to determine how changes in the cell boundary impact overall cell growth and patterning."
LobeFinder could also be used to identify causal relationships between cell shape changes over time and the chemical signals firing below the cell surface, Umulis said.
To use LobeFinder, a user feeds the x-y coordinates of a pavement cell boundary into the algorithm, which applies a convex hull to the image - like "putting a rubber band around it," Umulis said. LobeFinder identifies lobes by noting where the cell boundary meets the "rubber band" and examining whether the indentations on either side of that point are large enough to qualify it as a lobe.
While designed for pavement cells, the algorithm could measure changes in the boundaries of a number of irregularly shaped objects, such as immune cells on the move, shifts in the size of the Arctic ice cap or plumes of oil as they spread across the Gulf of Mexico, the researchers said.
Szymanski and Umulis envision LobeFinder as being one component in a cache of tools that together could provide automated phenotyping - the measurement of an organism's observable characteristics - of cellular and sub-cellular plant features.
"Phenotyping at this high resolution allows you to understand what dynamic cellular processes and behaviors can explain characteristics at the organ or tissue level," Szymanksi said. "Otherwise, you're blind to the mechanism."
The team is working on modifying LobeFinder to analyze cell shape changes across a population of cells.
The researchers are filing a disclosure agreement with the Office of Technology Commercialization.
The paper was published in Plant Physiology on June 10 and is available at http://www.plantphysiol.org/content/early/2016/06/10/pp.15.00972.full.pdf+html?sid=b544846f-fab2-4c1f-a714-9fdb5efd4d43.
The National Science Foundation provided funding for the research.
Writer: Natalie van Hoose, 765-496-2050, nvanhoos@purdue.edu
Sources: Daniel Szymanksi, 765-494-8092, dszyman@purdue.edu
David Umulis, 765-494-1233, dumulis@purdue.edu
ABSTRACT
LobeFinder: a convex hull-based method for quantitative boundary analyses of lobed plant cells
Tzu-Ching Wu 1; Samuel A. Belteton 2; Jessica Pack 1; Daniel B. Szymanski 2, 3, 4; David M. Umulis 1, 5
1 Department of Agricultural and Biological Engineering, 225 S. University Street, Purdue University, West Lafayette, IN 47907 USA
2 Department of Botany and Plant Pathology, 915 W. State Street, Purdue University, West Lafayette, IN 47907 USA
3 Department of Agronomy, 915 W. State Street, Purdue University, West Lafayette, IN 47907 USA
4 Department of Biological Sciences, 915 W. State Street, Purdue University, West Lafayette, IN 47907 USA
5 Weldon School of Biomedical Engineering, 206 S. Martin Jischke Drive, Purdue University, West Lafayette, IN 47907 USA
E-mail: dsysman@purdue.edu
Dicot leaves are comprised of a heterogeneous mosaic of jigsaw puzzle piece-shaped pavement cells that vary greatly in size and the complexity of their shape. Given the importance of the epidermis and this particular cell type for leaf expansion, there is a strong need to understand how pavement cells morph from a simple polyhedral shape into highly lobed and interdigitated cells. At present, it is still unclear how and when the patterns of lobing are initiated in pavement cells, and one major technological bottleneck to address the problem is the lack of a robust and objective methodology to identify and track lobing events during the transition from simple cell geometry to lobed cells. We developed a convex hull-based algorithm termed LobeFinder to identify lobes, quantify geometric properties, and create a useful graphical output for further analysis. The algorithm was validated against manually curated cell images of pavement cells of widely varying sizes and shapes. The ability to objectively count and detect new lobe initiation events provides an improved quantitative framework to analyze mutant phenotypes, detect symmetry-breaking event sin time-lapse image data, and quantify the time-dependent correlation between cell shape change and intracellular factors that may play a role in the morphogenesis process.
Agricultural Communications: (765) 494-2722;
Keith Robinson, robins89@purdue.edu
Agriculture News Page

