Tyrosine Kinases
The transduction of extracellular signals across the membrane to initiate complex cascades of molecular events regulates numerous vital cellular processes. The molecular basis of phospho-tyrosyl-regulation of Src-family and Syk-family protein tyrosine kinases in signaling pathways is a current focus of the lab.
  |
Src Kinase
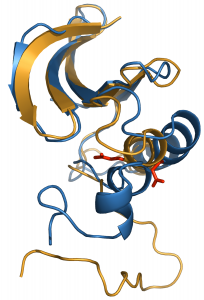
The conformational activation of Src kinase must be carefully regulated in cells for proper function and to avoid disease. The active and down-regulated forms of Src are defined by crystallography, but the transition between them is transient and only computer simulation can describe structures and energetics along the path between them. We have developed and applied computer simulation methods to investigate protein conformational transitions. An adaptively biased path optimization (ABPO) approach was developed to explore conformational transitions. ABPO investigations, along with other path search algorithms, discovered a switched electrostatic network involved in the transition. This insight from simulation suggests SEN as a mechanism for limiting the conformational space sampled between inactive and active forms of Src kinase. These investigations also established that the major energetic barrier for activation is the rotation of an α-helix known as helix C, which is conserved in structures across the human ‘kinome.’ That helix C is the energetic barrier explains a range of regulatory mechanisms involving highly diverse structures; that is, how kinase interactions observed crystallographically and validated by mutagenesis can regulate activation is rationalized by the energetic profile from the transition pathway calculations.
|
 |
Syk Kinase
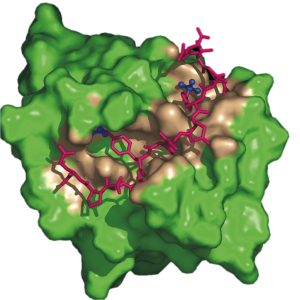
We are also actively investigating the regulation of Syk tyrosine kinase binding to membrane immune receptors and other signaling proteins. Syk recognizes doubly phosphorylated regions of the receptor called ITAM to initiate membrane signaling in an adaptive immune response. Syk additionally plays a role in signaling away from the membrane. Together, therefore, the interactions of Syk kinase must be orchestrated among many possible downstream binding partners. The overall goal is to understand how Syk tyrosine phosphorylation determines signaling outcome. We have looked at multiple complexes for Syk, including those with two phosphotyrosines binding to only one SH2 domain or binding to tandem SH2s.
|

