Gold nanoparticles help target, quantify breast cancer gene segments in a living cell
April 22, 2014
 |
|
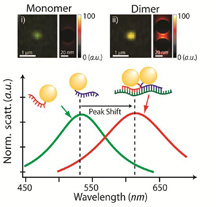
A single gold nanoparticle, or monomer, appears green when illuminated (top left), while a pair of gold nanoparticles bound to an mRNA splice variant, or dimer, appears reddish (top right). Monomers and dimers also scatter light differently, as shown in the graph above. (Purdue University image / Joseph Irudayaraj) |
WEST LAFAYETTE, Ind. - Purdue University researchers have developed a way to detect and measure cancer levels in a living cell by using tiny gold particles with tails of synthetic DNA.
A team led by Joseph Irudayaraj, professor of agricultural and biological engineering, used gold nanoparticles to target and bind to fragments of genetic material known as BRCA1 messenger RNA splice variants, which can indicate the presence and stage of breast cancer. The number of these mRNA splice variants in a cell can be determined by examining the specific signal that light produces when it interacts with the gold nanoparticles.
"This is a simple yet sophisticated technique that can be used to detect cancer in a single cell and determine how aggressive it is," said Irudayaraj, who is also the deputy director of the Bindley Bioscience Center. "Being able to quantify these genetic molecules could ultimately help clinicians provide better and more individualized treatment to cancer patients."
The technique also could increase our understanding of cell biology and paves the way for genetic profiling and diagnosis based on a single cell, Irudayaraj said.
BRCA1 is a tumor suppressor gene that can transform a cell into a cancerous type under certain circumstances. Measuring the number of BRCA1 mRNA splice variants in a cell can indicate if the gene is being under-expressed, a possible sign of breast cancer.
But current methods of detecting cancer rely on samples made up of hundreds or thousands of cells and cannot provide detailed information about how genes tied to cancer are being expressed in individual cells.
 |
|
Joseph Irudayaraj |
Irudayaraj and his team are the first to detect and quantify BRCA1 mRNA splice variants - fragments of genetic material that are removed when mRNA is formed - in a single cell. Splice variants can determine the fate of a cell and how specific proteins are expressed. Errors in the splicing process have been linked to a variety of diseases.
"With this method, we are basically able to spot a needle in a haystack - and we can determine if there are five needles in that haystack or if there are 50," he said.
Irudayaraj and his then-graduate research assistant, Kyuwan Lee, who is the first author of the study, adapted common nanotechnology methods to tackle the challenge of pinpointing mRNA splice variants in a living cell. They fabricated gold nanoparticles - more than 1,000 times smaller than the diameter of a human hair - and tagged them with strands of DNA complementary to BRCA1 mRNA splice variants.
When injected into a cell, the nanoparticles attached to either end of mRNA splice variants, forming structures known as dimers - each "like a couple holding hands," Irudayaraj said.
Because dimers give off a unique signal in the presence of light, the researchers could measure the number of dimers by illuminating the cell with a simple light source. The number of dimers corresponded to the number of BRCA1 mRNA splice variants in a cell.
Light behaves differently when it shines on a single gold particle, allowing the researchers to differentiate between dimers and free-floating gold particles.
The researchers used two methods to quantify the dimers: spectroscopy, which measures the way light scatters when it encounters an object, and a colorimetric image on which dimers show as reddish dots while single gold particles appear green.
The technique can quantify mRNA splice variants in a single cell in about 30 minutes.
Irudayaraj is modifying the system to speed up the process so that it can be used in tissue biopsies.
"If we can quantify key mRNA at single cell resolution in a tissue biopsy, that will be very powerful in terms of refining treatment protocols for key diseases," he said.
The paper was published in Nature Nanotechnology on April 20 and is available at http://www.nature.com/nnano/journal/vaop/ncurrent/full/nnano.2014.73.html.
The National Science Foundation, the Indiana Clinical Transitional Sciences Institute, Purdue Center for Cancer Research, Samsung and Stanford National Institute of Health funded the research.
Writer: Natalie van Hoose, 765-496-2050, nvanhoos@purdue.edu
Source: Joseph Irudayaraj, 765-494-0388, josephi@purdue.edu
ABSTRACT
Quantitative imaging of single mRNA splice variants in living cells
Kyuwan Lee 1, 2; Yi Cui 2; Luke P. Lee 1; Joseph Irudayaraj 2
1 Department of Bioengineering, Department of Electrical Engineering and Computer Science, University of California Berkeley, California 94720
2 Department of Agricultural and Biological Engineering, Bindley Bioscience Center, Purdue University, 225 South University Street, West Lafayette, Indiana 47907
E-mail: lplee@berkeley.edu, josephi@purdue.edu
Alternative messenger RNA (mRNA) splicing is a fundamental process of gene regulation, and errors in RNA splicing are known to be associated with a variety of different diseases. However, there is currently a lack of quantitative technologies for monitoring mRNA splice variants in cells. Here, we show that a combination of plasmonic dimer probes and hyperspectral imaging can be used to detect and quantify mRNA splice variants in living cells. The probes are made from gold nanoparticles functionalized with oligonucleotides and can hybridize to specific mRNA sequences, forming nanoparticle dimers that exhibit distinct spectral shifts due to plasmonic coupling. With this approach, we show that the spatial and temporal distribution of three selected
splice variants of the breast cancer susceptibility gene, BRCA1, can be monitored at single-copy resolution by measuring the hybridization dynamics of the nanoplasmonic dimers. Our study provides insights into RNA and its transport in living cells, which could improve our understanding of cellular protein complexes, pharmacogenomics, genetic diagnosis and gene therapies.
Ag Communications: (765) 494-2722;
Keith Robinson, robins89@purdue.edu
Agriculture News Page

