Findings reveal clues to functioning of mysterious 'mimivirus'
May 14, 2015
 |
|
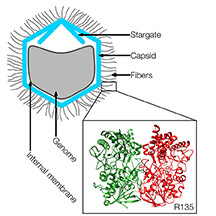
This graphic shows the location of a key protein called R135, contained in fibers on the outer surface of an unusually large virus called the mimivirus. Findings by Purdue researchers are aiding efforts to learn more about the mysterious virus, including its hosts and unknown functions (Klose et al./Structure 2015) |
WEST LAFAYETTE, Ind. - Researchers have discovered the structure of a key protein on the surface of an unusually large virus called the mimivirus, aiding efforts to determine its hosts and unknown functions.
The mimivirus was initially thought to be a bacterium because it is much larger than most viruses. It was isolated by French scientists in 1992 but wasn't confirmed to be a virus until 2003.
In the laboratory, the virus has been studied while infecting amoebas, but its natural hosts in the wild and many details about the virus remain unknown, said Michael Rossmann, Purdue University's Hanley Distinguished Professor of Biological Sciences.
He led a team of researchers who discovered the structure of a key, enzyme-like protein called R135, which is contained in fibers on the outer surface of the virus. The structure of R135 is similar to an enzyme called aryl alcohol oxidase, which is found in a fungus and is involved in biodegrading lignin in plant cell walls.
"This could tell us something about the mimivirus's natural hosts," Rossmann said. "We think there must be another host, something different than amoebas and that this enzyme helps the virus get into this host. Perhaps R135 participates in the degradation of lignin so that the virus can enter a host such as lignin-containing algae."
Also suggesting the possibility of alternative hosts is the recent discovery that mimivirus has been found to be abundant in oysters. Antibodies against mimivirus have been found in humans, and the virus has been discovered inside specialized cells in humans called macrophages, but whether it actually infects people is not known.
"I wouldn't say it infects humans, but it can propagate in humans because macrophages take up this virus and it can propagate in these," said Thomas Klose, a assistant research scientist at Purdue working with Rossmann. "Overall, there are many unknowns."
Research findings will appear in the June 2 issue of the journal Structure.
Mimiviruses are among the largest known viruses. The virus approaches the size of bacteria and is about half of a micron in diameter, more than 10 times larger than the virus that causes the common cold and large enough to be seen with a light microscope. Other viruses are too small to be seen with conventional light microscopes.
"It's one of the biggest viruses and has about a thousand genes, many of which have unknown functions, and it seems to be like a melting pot of enzymes," Klose said.
The R135 protein likely plays a key role in the virus's relationship with a smaller, "satellite virus" aptly called Sputnik.
Sputnik is referred to as a satellite virus because it can only be propagated in combination with another virus, in this case the mimivirus, which infects amoebas in laboratory research. While inside the single-cell amoeba, the DNA for both Sputnik and mimivirus are replicated, allowing the viruses to multiply.
"It is very likely that this R135 is the protein that Sputnik uses to co-infect with mimivirus," Rossmann said.
The Structure paper was authored by Klose; Dominik A. Herbst, a former intern and now a graduate student at the University of Basel in Switzerland; Purdue graduate students Hanyu Zhu and Joann P. Max in the Department of Chemistry; Hilkka I. Kenttämaa, a professor in the Department of Chemistry; and Rossmann.
The researchers used a technique called X-ray crystallography to determine the protein's structure.
The research was funded by the National Institutes of Health.
Writer: Emil Venere, 765-494-4709, venere@purdue.edu
Sources: Michael Rossmann, 765-494-4911, mr@purdue.edu
Thomas Klose, tklose@purdue.edu
Note to Journalists: A copy of the article is available by contacting Emil Venere, 765-494-4709, venere@purdue.edu
ABSTRACT
A Mimivirus enzyme that participates in viral entry
Thomas Klose,1 Dominik A. Herbst,1,2 Hanyu Zhu,3 Joann P. Max,3 Hilkka I. Kenttämaa3 and
Michael G. Rossmann1,*
1 Department of Biological Sciences, Purdue University
2 Current address: Biozentrum, University of Basel, Klingelbergstrasse
3 Department of Chemistry, Purdue University
* Correspondence: Michael G. Rossmann, Department of Biological Sciences, 240 S. Martin Jischke Drive, Purdue University, West Lafayette, IN 47907-2032, USA. Tel.:+1 765-494-4911; Fax: +1 765-496-1189; E-mail: mr@purdue.edu
Mimivirus was initially identified as a bacterium because its dense, 125nm long fibers stained Gram-positive. These fibers probably play a role during the infection of some host cells. The normal hosts of Mimivirus are unknown, but in the laboratory Mimivirus is usually propagated in amoeba. The structure of R135, a major component of the fibrous outer layer of Mimivirus, has been determined to 2Å resolution. The protein's structure is similar to members of the glucose-methanol-cholin oxidoreductase family, which have an N-terminal FAD binding domain and a C-terminal substrate recognition domain. The closest homologue to R135 is an aryl alcohol oxidase that participates in lignin biodegradation of plant cell walls. Thus R135 might participate in the degradation of their normal hosts, including some lignin-containing algae.

