InferNet
Gene Function Inference By Leveraging Large, Organ-Specific Expression Datasets And Validation Of Non-Redundant Regulators
This project combines computational approaches, such as machine learning, regulatory network inference and phylogenomics, with molecular approaches, such as metabolite profiling and ChIP-Seq, to characterize the role played by key transcription factors (TF) in regulating traits of agronomic or biofuel interest. For the majority of plant genes knowledge of their function, especially the association of gene to phenotype, remains stubbornly incomplete despite focused efforts by the plant research community. This project focuses on the biofuel trait of seed oil synthesis as a proof of concept for an approach developed that is extensible to any agronomic/biofuel trait of interest. For a given trait of interest, we predict novel TF regulators by leveraging the power of large expression data sets in a model species and then translate that knowledge to a target biofuel species for immediate benefit to the DOE mission. Two of the principal challenges in determining gene function are: 1. Subtle phenotypes that occur in organ and/or developmental stage specific manner and 2. Functional redundancy among gene family members masks phenotypic effects via genetic or dosage compensation. In this project, we will develop a general approach that addresses these two major concerns by focusing on regulation of a biological process of interest (e.g., lipid biosynthesis) in an organ specific manner (e.g., in seeds) and by estimating the likelihood of a given TF being redundant in its function (Aim 1). We will validate our functional predictions, using transgenic lines (Aim 2), via phenotypic assays (Aim 3a) and by identifying the specific target genes these TFs regulate (Aim 3b). Finally, we translate the validated TF regulation knowledge gained in a model species (Arabidopsis thaliana) to biofuel crops (e.g., Camelina sativa) (Aim 4).
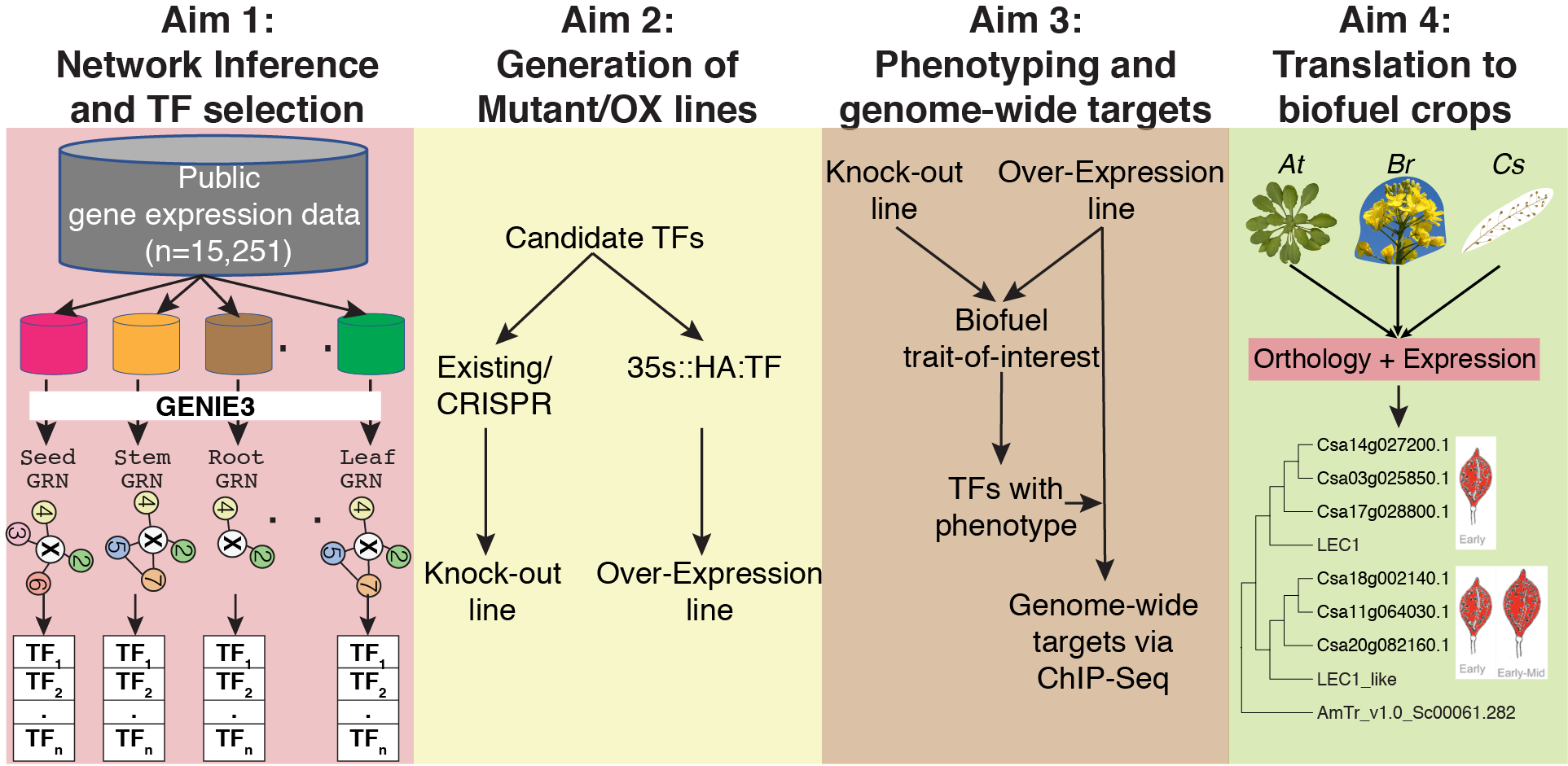
Outcome: This DOE funded project focuses on the seed lipid biosynthesis trait as a proof-of-concept and aims to identify multiple novel regulators of this trait. The data and algorithms developed in this project should prove valuable to any research group interested in discovering novel TF regulators of any plant process. This project provides the data and tools to discover such regulators in the model plant Arabidopsis as well as a computational pipeline to translate the knowledge gained in Arabidopsis to a biofuel species.
The Team:
Kranthi Varala (PI)
Ying Li (Co-PI)
Karen Hudson (Co-PI)