

March 11, 2008
Virus views enhanced using nation's largest Condor flock
WEST LAFAYETTE, Ind. -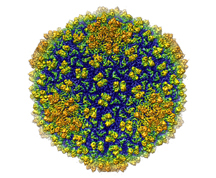 |
Wen Jiang, an assistant professor of biological sciences, led a team that used the emerging technique of single-particle electron cryomicroscopy to capture a three-dimensional image of a virus at a resolution of 4.5 angstroms. Approximately 1 million angstroms would equal the diameter of a human hair.
The breakthrough was enabled through the use of Purdue's Condor distributed computing grid, which comprises more than 7,000 computers. Purdue's Condor flock - which links desktop machines, computers in student computing labs, and powerful research computers - is the largest distributed computing network at a university.
Computer programs are used to extract the signal from the microscope and to combine thousands of two-dimensional images into an accurate three-dimensional image that maps the structure of the virus. This requires use of a large data set and could not have been done without the resources of Purdue's Office of Information Technology, or ITaP, Jiang said.
"ITaP provided us with computational power at the supercomputer scale that was necessary for this work," he said. "Purdue's Condor flock allowed us to take advantage of the power of 7,000 computers. This was a critical element to our success."
John Campbell, associate vice president of information technology, says this is one example of the type of discoveries that can be made using distributed computing.
"This work was done while the computers weren't otherwise being used," Campbell said. "We are capturing what would have been wasted computer cycles and putting them to use doing groundbreaking science in a cost-efficient way."
Jiang plans to continue to refine every step of the process to improve the capabilities of the technique and to examine more medically relevant virus species.
The imaging technique, called cryo-EM, has the added benefit of maintaining the sample being studied in a state very similar to its natural environment. Other imaging techniques used regularly, such as X-ray crystallography, require the sample be manipulated.
"This method offers a new approach for modeling the structure of proteins in other macromolecular assemblies, such as DNA, at near-native states," Jiang said. "The sample is purified in a solution that is very similar to the environment that would be found in a host cell. It is as if the virus is frozen in glass and it is alive and infectious while we examine it."
A paper detailing the work was published in the Feb. 28 issue of Nature.
Cryo-EM requires high-end electron microscopes and powerful computing resources. The research team used the Baylor College of Medicine's cryoelectron microscope. It is expected that Purdue will install a state-of-the-art cryoelectron microscope in 2009.
In 2006 Purdue received a $2 million grant from the National Institute of Health to purchase the microscope. It will be installed in Hockmeyer Hall of Structural Biology, expected to open in 2009.
In addition to Jiang, Matthew L. Baker, Joanita Jakana and Wah Chiu from Baylor College of Medicine, and Peter R. Weigele and Jonathan King from Massachusetts Institute of Technology worked on the project, which was funded by the National Institutes of Health and the National Science Foundation.
Writers: Elizabeth K. Gardner, (765) 494-2081, ekgardner@purdue.edu
Steve Tally, (765) 494-9809, tally@purdue.edu
Source: Wen Jiang, (765) 496-8436, jiang12@purdue.edu
John Campbell, (765) 494-1289. john-campbell@purdue.edu
Purdue News Service: (765) 494-2096; purduenews@purdue.edu
IMAGE CAPTION:
Shown is an image of bacteriophage Epsilon15 studied by Wen Jiang, an assistant professor of biological sciences at Purdue. The bacteriophage is shown at a resolution of 4.5 angstrom - the highest resolution achieved for a living organism of this size. (Graphic/Wen Jiang lab)
A publication-quality image is available at https://www.purdue.edu/uns/images/+2008/jiang-bacteriophage.jpg
To the News Service home page
If you have trouble accessing this page because of a disability, please contact Purdue News Service at purduenews@purdue.edu.
