
| RELATED INFO |
| * Purdue Department of Agronomy |
| * Proceedings of the National Academy of Sciences |

March 6, 2008
Newly defined signaling pathway could mean better biofuel sources
WEST LAFAYETTE, Ind. - |
The pathway moves materials that determine cell shape and size through a system of signaling proteins, said Dan Szymanski, a plant geneticist and cellular biologist. By learning more about the growth and development process, it may be possible to engineer plants with improved properties such as cell walls that are more massive or are more easily fermented in the biofuel process.
"We expect that cell wall material will be a major source of biomass from plants designated for biofuel production," Szymanski said. "We need to learn more about how plant cells control the quality and amount of cell wall material."
He and his research team investigated plant growth and cell wall development from several scientific approaches in determining the cascade of events that leads to changes in the cell wall. They discovered that a protein called "SPIKE1" directs the protein signaling pathway. They report their findings in "Early Edition," the online publication of the journal Proceedings of the National Academy of Sciences. The study also will be published in the journal's March 11 print issue.
"Plant cells grow by expansion, which is cell wall synthesis coupled with an increase in cell size," Szymanski said. "The key questions we need to answer in trying to create plants more valuable for biofuel production center on understanding how plants integrate metabolism, cell growth and biomass production."
To answer those questions and be able to engineer plants for improved growth of biomass for alternative fuels, Szymanski and other scientists must investigate molecular function.
 |
"Our research is focused on understanding signaling mechanisms," he said. "How does a cell interpret multiple types of information and then translate that information to a signal that says, 'Grow here, or modify or reinforce the cell wall here.' Or how does a cell know to make new cytoskeleton filaments at a certain time and place to define regions of growth that determine the cell's shape and size?"
Actin filaments comprise the cytoskeleton, which is the roadway for delivery and recycling of materials that drive plant growth and determine the cell shape and size. Actin is an abundant protein in organisms that have multiple cells with nuclei.
SPIKE1 is a master regulator of many growth control pathways, including the protein signaling pathway that produces the cytoskeleton. Szymanski and his colleagues were able to demonstrate that one of SPIKE1's functions is to control production of actin filament, which defines localized cell regions for delivery and recycling of growth materials.
"Wall construction in plants, just as in a road project, is a coordinated effort," Szymanski said. "The supply and demand of the materials needed for growth must be coordinated. The question is, how do cells regulate this?"
The signaling pathway, headed by SPIKE1, is responsible for organizing activities during construction - delivering materials and recycling materials that are used during growth, he said. After SPIKE1 initiates communication among proteins along the pathway, actin filaments are produced and changes in cell shape and size occur.
Cells also must coordinate with the activities of surrounding cells that have different shapes and functions.
"Cell expansion occurs in a crowded, but accommodating environment," Szymanski said. "As neighboring cells expand, this growth intrudes upon a neighbor. SPIKE1 generates signals so that cells can coordinate with neighboring cells' activities to promote organized cell expansion and proper cell-to-cell adhesion."
Szymanski and his colleagues used an altered version of the mustard family laboratory plant Arabidopsis to study SPIKE1's function and find the proteins that it activates and to which it binds.
They found that when they created mutant plants by switching off the SPIKE1 gene so that the function is lost, one result was improper growth that manifested as holes in the leaf epidermis.
By studying the results of turning off various other protein complexes in the pathway, Szymanski's team was able to follow the sequence of events that occur during signaling.
They also found that plants in which the function of one of the pathway's signaling proteins was altered resulted in mutants that all looked alike, Szymanski said. This suggested that the three major protein complexes the scientists investigated all function in a common pathway. The Purdue research team confirmed this by making double mutants - plants in which two of the proteins had been switched off. One of the pathway's protein complexes, called "WAVE," functions the same way in both humans and Arabidopsis, and the SPIKE1 signaling pathway is likely to function in other plants including rice and corn.
However, in other organisms with SPIKE1-like genes, switching off the gene kills the organism. This lethality has made it difficult for scientists to understand the function of SPIKE1 and comparable genes in other organisms, including humans. Since Arabidopsis survives when SPIKE1 is disrupted, the Purdue team was able to determine the signaling pathway.
The scientists hypothesize that SPIKE1 may both generate and organize protein complex signaling, Szymanski said. They also need to discover what activates SPIKE1. When the researchers understand enough about the processes involved in plant cell growth and development, then they may be able to design plants that are bigger with more cell wall that can be processed into biofuel.
"Learning more about SPIKE1 likely will help us gain a better understanding of the mechanics and regulation involved with the pathways that control cell architecture and development in plants, and also may be relevant to animal and human growth and development," Szymanski said.
The other researchers involved with this study were graduate student Dipanwita Basu, postdoctoral students Jie Le and Taya Zakharova, and research technician Eileen Mallery. All are in the Purdue Department of Agronomy.
The National Science Foundation and the Purdue Agricultural Research Program funded this project.
Writer: Susan A. Steeves, (765) 496-7481, ssteeves@purdue.edu
Source: Dan Szymanski, (765) 494-8092, dszyman@purdue.edu
Ag Communications: (765) 494-2722;
Beth Forbes, forbes@purdue.edu
Agriculture News Page
PHOTO CAPTION:
Plant geneticist Dan Szymanski of the Purdue Department of Agronomy is leading a research team that has defined a biochemical pathway that may make it possible to engineer plants with improved properties, potentially leading to better biofuel sources. The scientists are particularly interested in the signaling that leads to cell growth and development. (Purdue Agricultural Communication photo/Tom Campbell)
A publication-quality photo is available at https://www.purdue.edu/uns/images/+2008/Szymanski-Dan.jpg
PHOTO CAPTION:
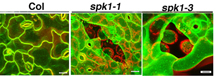
A Purdue research team is studying plant growth and cell wall development. By investigating plant cells at the molecular level, they may be able to design plants that are better sources of alternative transportation fuels. In these three slides, green outlines the outer epidermal cells. The red is from chloroplasts from the underlying cell layer. The final slide shows cells of a mutant plant in which a gene called SPIKE1 has been turned off. These mutant cells form abnormally and the cell walls won't properly adhere to each, resulting in holes in the epidermis that you can see through. (Photo courtesy of Dan Szymanski, Purdue University)
A publication-quality photo is available at https://www.purdue.edu/uns/images/+2008/szymanski-biofuelSPIKE1.jpg
A SPIKE1 Signaling Complex Controls Actin-Dependent
Cell Morphogenesis through the Heteromeric WAVE and ARP2/3 Complexes
Dipanwita Basu, Jie Le, Taya Zakharova, Eileen L. Mallery, and Daniel B. Szymanski
During morphogenesis, the actin cytoskeleton mediates cell-shape change in response to growth signals. In plants, actin filaments organize the cytoplasm in regions of polarized growth, and the filamentous arrays can be highly dynamic. Small GTPase signaling proteins termed Rho of plants (ROP)/RAC control actin polymerization. ROPs cycle between inactive GDP-bound and active GTP-bound forms, and it is the active form that interacts with effector proteins to mediate cytoskeletal rearrangement and cell-shape change. A class of proteins termed guanine nucleotide exchange factors (GEFs) generates GTP–ROP and positively regulates ROP signaling. However, in almost all experimental systems, it has proven difficult to unravel the complex signaling pathways from GEFs to the proteins that nucleate actin filaments. In this article, we show that the DOCK family protein SPIKE1 (SPK1) is a GEF, and that one function of SPK1 is to control actin polymerization via two heteromeric complexes termed WAVE and actin-related protein (ARP) 2/3. The genetic pathway was constructed by using a combination of highly informative spk1 alleles and detailed analyses of spk1, wave, and arp2/3 single and double mutants. Remarkably, we find that in addition to providing GEF activity, SPK1 associates with WAVE complex proteins and may spatially organize signaling. Our results describe a unique regulatory scheme for ARP2/3 regulation in cells, one that can be tested for widespread use in other multicellular organisms.
To the News Service home page
If you have trouble accessing this page because of a disability, please contact Purdue News Service at purduenews@purdue.edu.
