
May 2, 2005
Findings reinforce theories about how viruses evolve
WEST LAFAYETTE, Ind. –Two viruses that split from a common ancestor possibly a billion years ago still have the same protein "fold" in their outer shells, shedding light on how viruses evolved, Purdue University researchers have found.
|
The outer shells – or capsids – of viruses contain proteins, which are made of a string of building blocks called amino acids. Proteins fold into specific shapes, depending on the sequence of amino acids.
The team of structural biologists has determined that the phi 29 virus, which attacks the soil bacterium bacillus subtilis, possesses the same protein fold in its capsid as the fold found in the HK97 virus, which infects E. coli bacteria.
"These findings are made possible by studying the capsid structure in nearly atomic-scale detail," said Marc C. Morais, a postdoctoral researcher in the laboratory of Michael Rossmann, the Hanley Distinguished Professor of Biological Sciences in Purdue’s College of Science.
The researchers used a powerful imaging tool called cryo-electron microscopy to determine the three-dimensional structure of phi 29 down to a resolution of 7.9 angstroms. An angstrom is one ten-billionth of a meter, or roughly one-millionth as wide as a human hair.
"What we're seeing is helping to confirm ideas about how viruses evolve," Morais said. "Other researchers previously discovered that seemingly unrelated viruses that infect mammals use the same capsid fold. Our new findings point to a similar phenomenon in bacteriophages, or viruses that infect bacteria."
A research paper about the findings appeared in April as the cover story of the journal Molecular Cell, vol. 18, no. 2. The paper was written by Morais, Purdue postdoctoral fellow Kyung H. Choi, Purdue electron microscopist Paul R. Chipman, Rossmann, and University of Minnesota researchers Dwight L. Anderson and Jaya S. Koti.
Other scientists recently found that another bacteriophage, called P22, contains the same capsid fold as HK97. Now the Purdue team has discovered the same fold in phi 29, leading to the likelihood that the fold is common among bacteriophages.
Phi 29 is one of a group of viruses that have double-stranded DNA, rather than a single strand as many others do. Phi 29's relative simplicity and small size make it an attractive choice for researchers interested in investigating viral structure.
Morais said the findings are particularly interesting because the two viruses attack different organisms yet are similar in the structure and organization of their capsid proteins.
Using cryo-electron microscopy, specimens are first frozen before they are studied with an electron microscope. The technique yields more accurate three-dimensional images than conventional methods in which specimens are dried and stained because ice is closer to a natural environment.
The technique enables scientists to study objects in details as small as about 8 angstroms, which is required to see the shape of the protein fold.
"Hopefully, at some point somebody will extract principles of how the pieces assemble," Morais said. "Considering the abundance of this type of virus, it is likely that the HK97 fold is one of the world's most common viral building blocks.
"These viruses probably represent the largest biomass on the entire planet. For every milliliter of seawater there is a huge number of phage that probably use the same fold as these. Their population is estimated to be 10 to the 31st power, which is a 1 with 31 zeroes after it – an inconceivably large number."
While phi 29's capsid has a prolate shape, giving it an appearance resembling a cylinder with hemispherical ends, HK97 is icosahedral, a shape with 20 sides. But despite differences in their overall shape, the two viruses are similar in the structure of their capsid proteins.
"It's important to note that HK97 infects E. coli, which is a so-called gram-negative bacterium, and phi 29 infects B. subtilis, which is called a gram-positive bacterium," Morais said. "These bacteria split into two types from a common ancestor about a billion years ago, which means these viruses also evolved that long ago."
The similarities between the two viruses suggest that many viruses may construct their capsids from a comparatively small number of building blocks and have done so for millions of years, Morais said.
"The same general assembly strategy that phi 29 uses is also used by certain animal viruses, such as herpes," he said. "Insights gained into the assembly of phi 29 may be applicable to mammalian viruses as well."
The researchers supported their findings about the shape of phi 29's fold with data from other researchers who determined HK97's fold using X-ray crystallography, another imaging technique.
The research was funded in part by the National Science Foundation and the National Institutes of Health.
Writer: Emil Venere, (765) 494-4709, venere@purdue.edu
Sources: Michael Rossmann, (765) 494-4911, mgr@indiana.bio.purdue.edu
Marc Morais, (765) 494-4910, morais@bilbo.bio.purdue.edu
Purdue News Service: (765) 494-2096; purduenews@purdue.edu
Note to Journalists: An electronic or hard copy of the research paper is available from Emil Venere, (765) 494-4709, venere@purdue.edu.
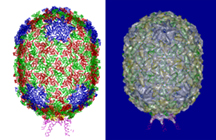
IMAGE CAPTION:
These two images depict the outer shell, or "capsid," of the phi 29 virus, which attacks the soil bacterium bacillus subtilis. The figure on the left shows the protein structure of the capsid, and the figure on the right overlays that structure on an "electron density map" of the outer shell created with a powerful imaging technique called cryo-electron microscopy. Purdue University researchers used the technique to determine the capsid's three-dimensional structure at nearly atomic-scale resolution. The scientists discovered that phi 29's capsid contains the same "fold" as another virus, called HK97, both of which split from a common ancestor possibly a billion years ago. The findings help to reinforce theories about how viruses have evolved over millions of years. (Images courtesy of Purdue University Department of Biological Sciences)
Conservation of the capsid structure in tailed dsDNA bacteriophages: the pseudo-atomic structure of phi 29
Bacteriophage phi 29 is one of the smallest and simplest known dsDNA phages, making it amenable to structural investigations. The three-dimensional structure of a fiberless, isometric variant has been determined to 7.9 Å resolution by cryo-electron microscopy (cryo-EM), allowing the identification of œ-helices and ß-sheets. Their arrangement indicates that the folds of the phi 29 and bacteriophage HK97 capsid proteins are similar, except for an additional immunoglobulin-like domain of the phi 29 protein. An atomic model that incorporates these two domains fits well into the croy-EM density of the T=3, fiberless isometric phi 29 particle, and cryo-EM structures of fibered isometric and fiberless prolate prohead phi 29 particles at resolutions of 8.7 Å and 12.7 Å, respectively. Thus, phi 29 joins the growing number of phages that utilize the HK97 capsid structure, suggesting that this protein fold may be as prevalent in capsids of dsDNA as is the jelly-roll fold in eukaryotic viruses.
To the News Service home page