Epigenetics
Histone modifications (ChIP-seq)
- Identification of histone marks on the genome
- Define chromatin states of the genome
- Integration of ChIP-seq data with RNA-seq data
MeDIP-Seq
- Identification and characterization of methylated regions on the genome
- Identification of differentially methylated regions (DMR)
Bisulfite Sequence analysis
- Generating single-base resolution DNA methylome
- Identification of differentially methylated regions (DMR)
- Integrate DMRs with transcriptomics
MNase-seq
- Map nucleosomes and other DNA-binding proteins
- Identification of location of various regulatory proteins in the genome
Samples of data visualization from our analyses
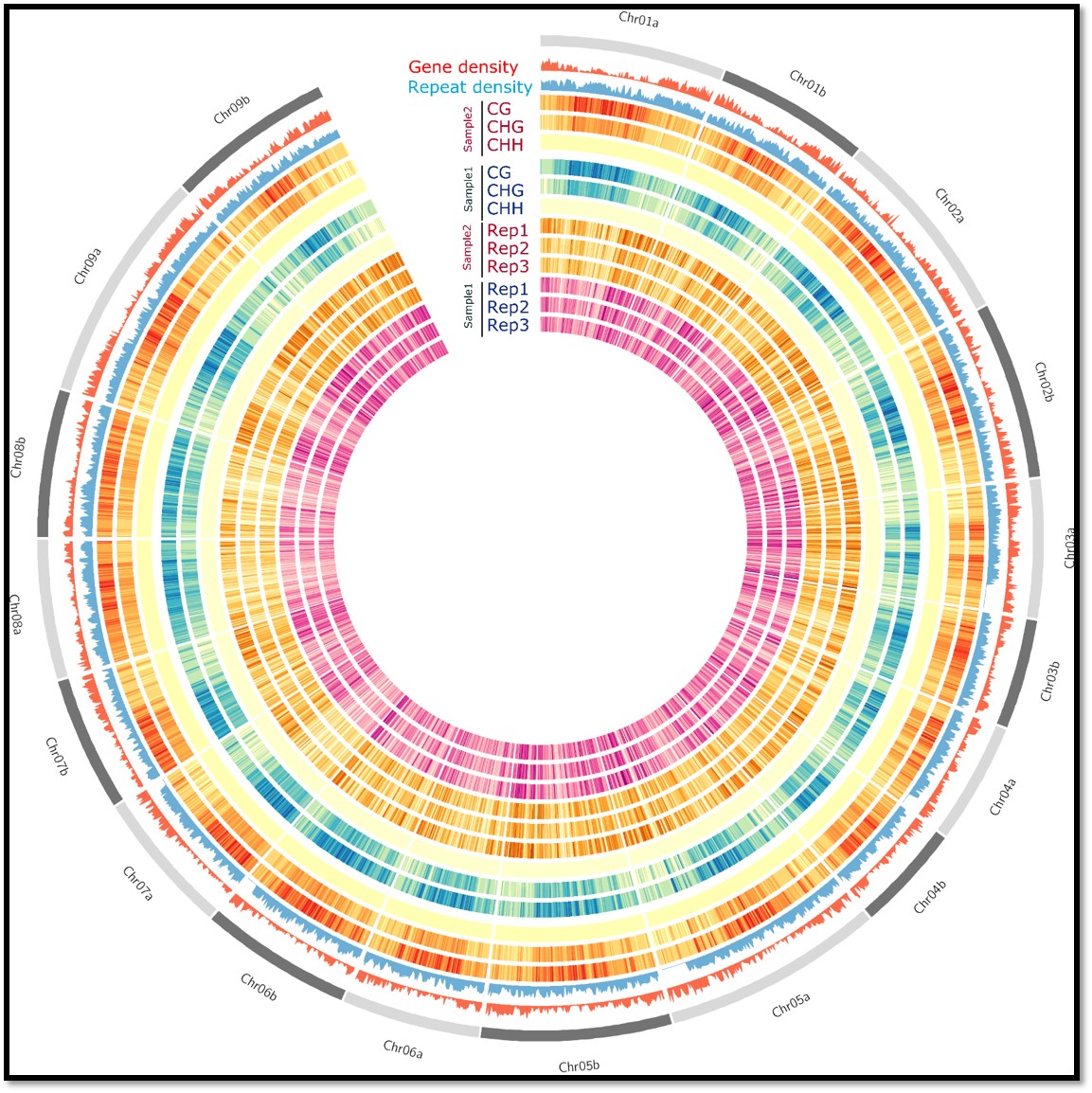
Circos plot of gene density, TE density, and DNA methylation across each chromosome
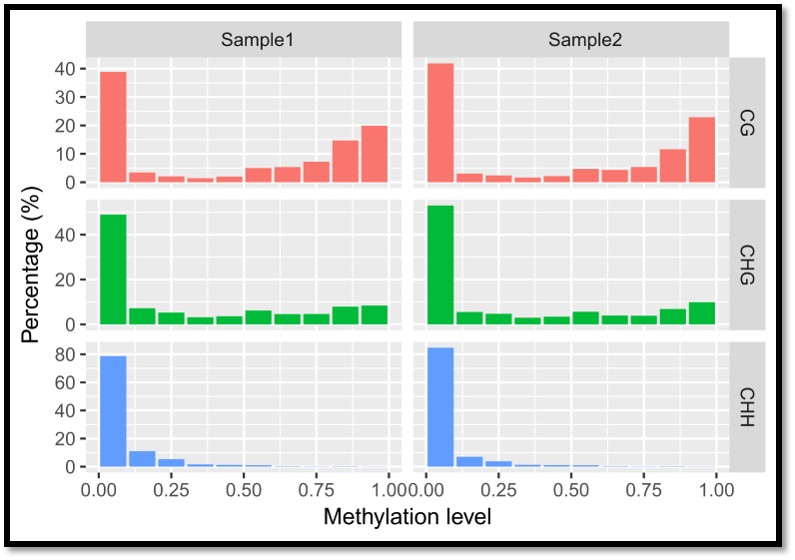
Distribution of DNA methylation for CG, CHG, and CHH contexts across genome
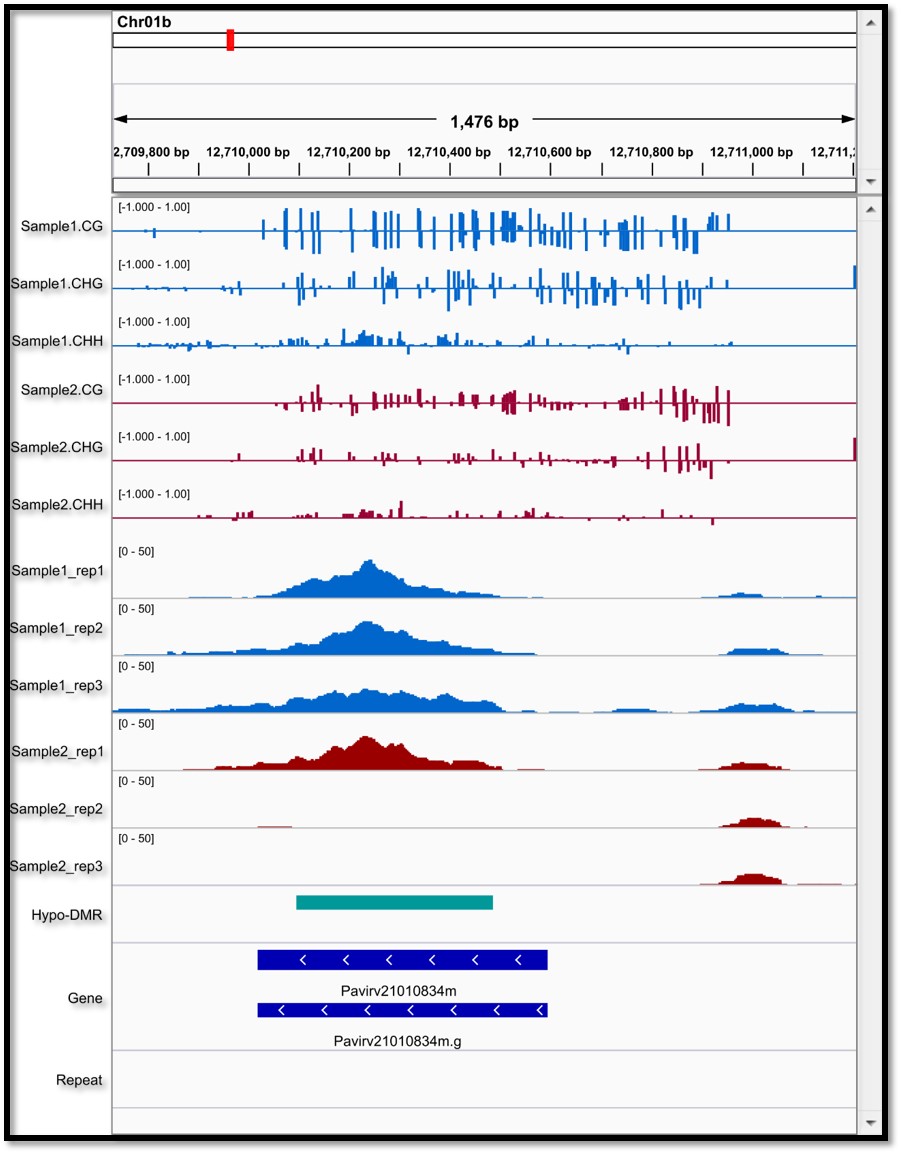
An IGV screenshot of DNA methylome
