October 26, 2022
Research into ancient lineage of microscopic fungi upends assumptions about its genetic relationships
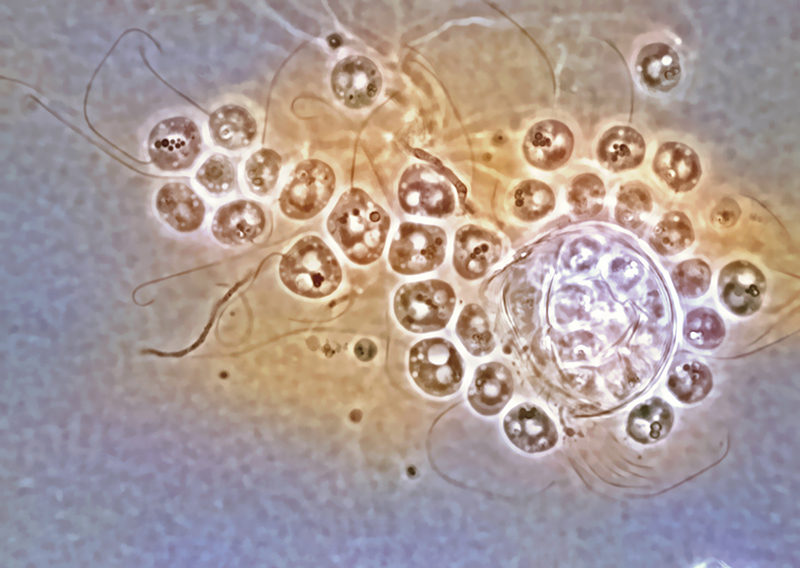
Rabern Simmons, Purdue’s new curator of fungi, named the Clydaea vesicula species of microscopic chytrid fungi after his mother, Clyda Rae Simms, a former teacher in Virginia’s Wise County school system, in 2009. A color overlay on the original black-and-white image enhances the specimen’s features. Credit: Rabern Simmons.
WEST LAFAYETTE, Ind. — Mycologists tend to base their evolutionary assumptions about all fungi on the higher fungi such as mushrooms, bread molds and yeasts. But that is a mistake, according to a major recent study published in the Proceedings of the National Academy of Sciences.
“The traits that the higher fungi possess are not indicative of the lower fungi, the early diverging fungi,” said Rabern Simmons, curator of fungi at the Purdue University Herbaria in Botany and Plant Pathology in the College of Agriculture.
The evolutionary history of the often-overlooked lineage of chytrid (pronounced kit-trid) fungi has vexed scientists for decades. The PNAS study has begun to clarify the complicated details of this lineage, which diverged from the common ancestor that it shares with animals about 750 million to 1 billion years ago.
“It takes a lot of our assumptions about early-diverging fungi and the increasing complexity of fungi as you work up the tree and throws them out the window,” Simmons said. “We showed that the chytrids still possess a lot of features that link them to that common ancestor.
In recent years, certain chytrid fungi have become a scourge of biodiversity. One infamous species of chytrids, described by Simmons’ graduate advisor and PNAS paper co-author Joyce Longcore at the University of Maine, has caused massive amphibian die-offs and extinctions.
Key to the PNAS study was how fungi that use different reproductive strategies were related to each other. Haploid organisms reproduce via mitosis cell division and have one set of chromosomes.
Diploid organisms have two sets of chromosomes, one from each parent, and most commonly reproduce via meiosis. This produces two haploid gametes, such as sperm and egg in humans, which fuse to form a new diploid organism.
“We started to look at the haploid versus diploid relationships in these fungi as opposed to higher fungi like mushrooms, bread molds and yeasts, things that people more commonly associate when they think of fungi,” Simmons said. “We found that a lot of the primary assumptions that haploid gives rise to diploid lifestyle — increasing complexity through the fungal kingdom — were not true. These things were reproducing by mitosis, but they weren’t always haploid; some were diploid.
Mycologists theorized that the higher fungi began as haploids that eventually gave rise to diploids.
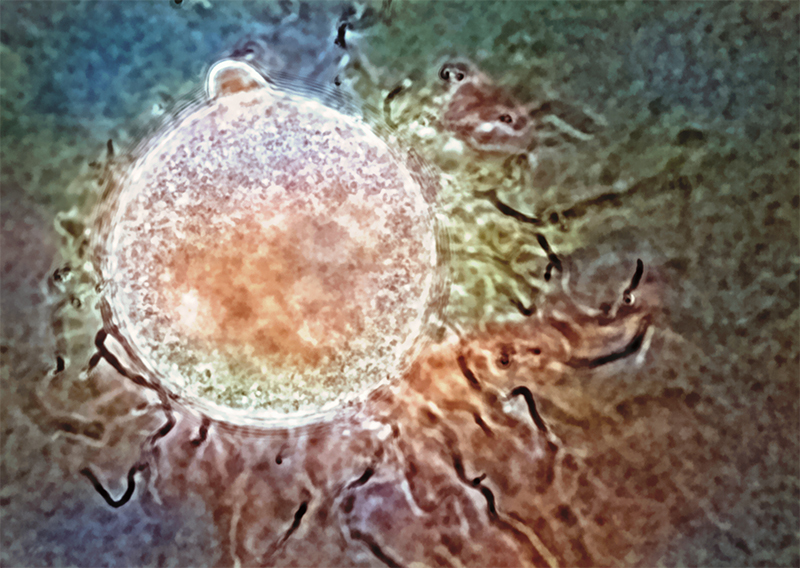 Fimicolochytrium jonesii. A color overlay on the original black-and-white image enhances the specimen’s features. Credit: Rabern Simmons.
Download image
Fimicolochytrium jonesii. A color overlay on the original black-and-white image enhances the specimen’s features. Credit: Rabern Simmons.
Download image
“They thought the chytrids probably were much the same. It turns out based on this genomic analysis that that’s not the case,” Simmons said.
The researchers concluded that fungal evolution proceeded more gradually and with more diversity than previously suspected. Their work led them to agree with some new classifications of chytrid fungi. Biologists classify humans, for example, as belonging to the phylum Chordata (backboned animals), the order of primates (which includes apes and monkeys), and the genus Homo.
As recently as the early 2000s, mycologists recognized five orders of chytrid fungi. The PNAS paper confirmed a dramatic reshuffling.
“Of those five orders that we understood to be chytrid, three are now their own phylum. And some genera within the remaining two have been pulled out and are now their own phyla. We understand a lot more about what’s going on,” Simmons said.
The PNAS paper relies heavily on the University of Michigan’s fungi culture collection that Simmons established before coming to Purdue earlier this year. About half of the 1,200 fungi isolates that the Michigan collection comprises came from Joyce Longcore’s laboratory at the University of Maine.
The co-authors also included a team of scientists at the U.S. Department of Energy’s Joint Genome Institute at the Lawrence Berkeley National Laboratory in California, who generated genome sequences for 69 chytrid fungi.
“When Joyce started collecting chytrids, I think she had little idea of just how instrumental a role they would play in resolving some of the big questions in fungal evolution, from the origins of life cycles to abilities to break down plant matter, to helping solve the mystery of the amphibian pandemic,” said the University of Michigan’s Timothy James, who led the PNAS study.
“It was a great pleasure working with Rabern, who was a great bridge between the traditional microscopy approaches and the modern genomics methods.”
Two leading chytrid fungi experts of the 20th century’s microscopy era were Purdue’s John Karling and the University of Michigan’s Frederick Sparrow. The new PNAS paper builds on Karling’s and Sparrow’s work.
“They had some things right and some things wrong,” Simmons said. “Hopefully, we can take the best of what they did, the best of what we’re doing, and further synthesize that into some excellent mycology that we can pass along to people that don’t even think of chytrid fungi when they think of fungi.”
Longcore commented on how the long-term interplay between Purdue and the University of Michigan regarding chytrid fungi has turned out.
"Sparrow and Karling were not close,” said Longcore, who worked for Sparrow. “And now Tim James has this great lab at the University of Michigan where Sparrow wrote this big monograph that includes the chytrids. And now Rabern’s at Purdue, and I hope he’ll have some time to work on chytrids. It’s a neat connection between Michigan and Purdue.”
Writer: Steve Koppes
Media contact: Maureen Manier, mmanier@purdue.edu
Source: Rabern Simmons, simmonddr@purdue.du
Agricultural Communications: 765-494-8415;
Maureen Manier, Department Head, mmanier@purdue.edu

